Principal Investigator
Prof. Dr. Norbert Tautz
Universität zu Lübeck
B3
PhD candidate
Sabine Höppner

B3
Project Summary
Topological and functional characterization of the pestiviral non-structural protein 4B (NS4B)
Pestiviruses such as bovine viral diarrhea virus (BVDV) are animal pathogens of the family Flaviviridae and are closely related to Hepatitis C virus (HCV). The viral particles are enveloped nucleocapsids that enclose a positive-sense, single-stranded RNA genome. The pestiviral genome is cap-independently translated into one polyprotein, which becomes co- and posttranslationally processed into mature proteins by cellular and viral proteases. The viral proteins can be distinguished into four structural (S) and eight non-structural (NS) proteins involved in diverse processes during the pestiviral life cycle like poly- protein processing, RNA replication, or virion morphogenesis. The limited repertoire of proteins encoded by the pestiviral genome makes it crucial to use intermediate cleavage products and distinct protein-protein interactions for the spatial and temporal regulation of these processes [1, 2]. Functional pestiviral RNA replication requires an interplay of NS3 to NS5B and a yet undefined set of cellular proteins (minimal replicase) [3]. While the function of certain replicase proteins like NS3 and NS4A was extensively studied, the pivotal role of NS4B for viral replication is not understood so far [4, 5, 6].
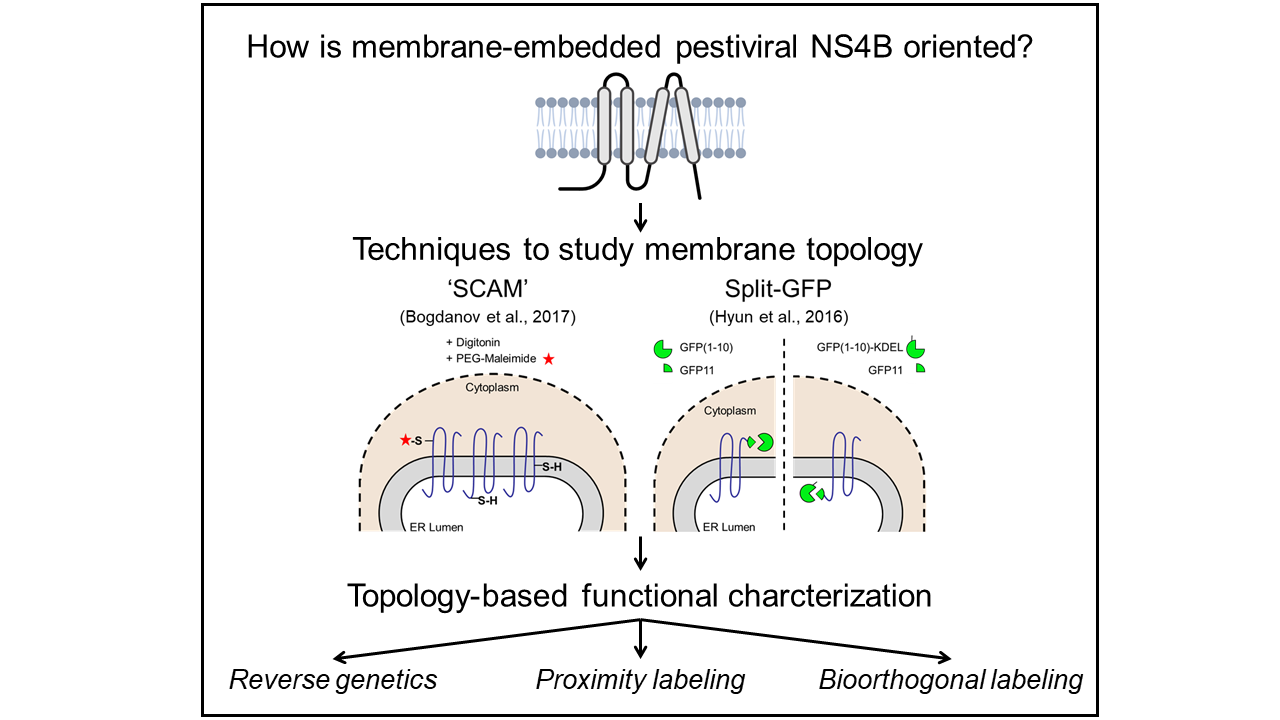
Since pestiviruses replicate and assemble at ER membranes, NS4B is assumed to anchor the viral replicase complex to these membrane structures as already suggested for Dengue virus [7, 8]. The membrane topology of NS4B is critical for functional protein-protein interactions; thus detailed knowledge of its topology represents a prerequisite to identify the interactions of this enigmatic protein. Topology studies of the full-length NS4B will be performed using ‘Substituted Cysteine Accessibility Method’ (SCAM) that utilizes the chemical reactivity of accessible thiol groups of cysteines to covalently modify the protein of interest with a membrane-impermeable maleimide compound after selective membrane permeabilization of the plasma membrane [9]. Afterward, the SCAM-based NS4B membrane topology model will be validated using a Split-GFP assay. Thereby, GFP is separated into a larger detector molecule expressed in the cytoplasm or the ER lumen, respectively, and the smaller sensor molecule becomes attached to C-terminally truncated NS4B mutants. In this assay, fluorescence will only be detectable if detector and sensor molecules reside in the same cellular compartment, hence enabling the determination of the orientation of the artificial NS4B C-terminus across the ER membrane [10]. The established BVDV NS4B membrane topology model will guide further functional characterization of this protein using reverse genetics, proximity labeling with NS4B-TurboID fusion proteins [11], and bioorthogonal labeling using UV-crosslinking of non-canonical amino acids [12].
B3
References
- Tautz, N., Tews, B.A., and Meyers, G. (2015). The Molecular Biology of Pestiviruses. Adv Virus Res 93, 47–160. https://doi.org/10.1016/bs.aivir.2015.03.002.
- Simmonds, P., Becher, P., Bukh, J., Gould, E.A., Meyers, G., Monath, T., Muerhoff, S., Pletnev, A., Rico-Hesse, R., Smith, D.B., et al. (2017). ICTV Virus Taxonomy Profile: Flaviviridae. Journal of General Virology 98, 2–3. https://doi.org/10.1099/jgv.0.000672.
- Behrens, S.E., Grassmann, C.W., Thiel, H.J., Meyers, G., and Tautz, N. (1998). Characterization of an autonomous subgenomic pestivirus RNA replicon. J Virol 72, 2364–2372. https://doi.org/10.1128/JVI.72.3.2364-2372.1998.
- Grassmann, C.W., Isken, O., and Behrens, S.E. (1999). Assignment of the multifunctional NS3 protein of bovine viral diarrhea virus during RNA replication: an in vivo and in vitro study. J Virol 73, 9196–9205. https://doi.org/10.1128/JVI.73.11.9196-9205.1999.
- Dubrau, D., Tortorici, M.A., Rey, F.A., and Tautz, N. (2017). A positive-strand RNA virus uses alternative protein-protein interactions within a viral protease/cofactor complex to switch between RNA replication and virion morphogenesis. PLoS Pathog 13, e1006134. https://doi.org/10.1371/journal.ppat.1006134.
- Grassmann, C.W., Isken, O., Tautz, N., and Behrens, S.E. (2001). Genetic analysis of the pes-tivirus nonstructural coding region: defects in the NS5A unit can be complemented in trans. J Virol 75, 7791–7802. https://doi.org/10.1128/jvi.75.17.7791-7802.2001.
- Murray, C.L., Jones, C.T., and Rice, C.M. (2008a). Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat Rev Microbiol 6, 699–708. https://doi.org/10.1038/nrmicro1928.
- Miller, S., Sparacio, S., & Bartenschlager, R. (2006). Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. The Journal of biological chemistry, 281(13), 8854–8863. https://doi.org/10.1074/jbc.M512697200.
- Bogdanov, M. (2017). Mapping of Membrane Protein Topology by Substituted Cysteine Accessibility Method (SCAM™). In: Journet, L., Cascales, E. (eds) Bacterial Protein Secretion Systems. Methods in Molecular Biology, vol 1615. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7033-9_9.
- Hyun, S.-I., Maruri-Avidal, L., and Moss, B. (2015). Topology of Endoplasmic Reticulum-Associated Cellular and Viral Proteins Determined with Split-GFP: Topology of ER-Associated Proteins. Traffic 16, 787–795. https://doi.org/10.1111/tra.12281.
- Cho, K. F., Branon, T. C., Udeshi, N. D., Myers, S. A., Carr, S. A., & Ting, A. Y. (2020). Proximity labeling in mammalian cells with TurboID and split-TurboID. Nature Protocols 2020 15:12, 15(12), 3971–3999. https://doi.org/10.1038/s41596-020-0399-0.
- Hermanson, G. T. (2013). The Reactions of Bioconjugation. In Bioconjugate Techniques (pp. 229–258). Elsevier. https://doi.org/10.1016/b978-0-12-382239-0.00003-0.
