Principal Investigator
Prof. Dr. Lars Redecke & Prof. Dr. Thomas Schulz
& Prof. Dr. Melanie Brinkmann
Universität zu Lübeck
& Hannover Medical School
& Technische Universität Braunschweig
C1
PhD candidate
Fatama Sornaly



C1
Project Summary
Structural and functional features of a conserved DNA-binding domain in herpes viral proteins as a basis for innovative targeted therapies

Human herpesviruses establish lifelong persistent infections that can play a key role in tumorigenesis and provide the basis for severe disease resulting from viral reactivation. Herpesviral proteins interacting with viral DNA (vDBPs) are key players in latency establishment, maintenance, and regulation. In the herpesviral replication cycle, vDBPs can be classified as latent or immediate-early (IE) lytic proteins. Latent proteins such as KSHV LANA-1 and EBV EBNA-1 of the gamma herpesvirus subfamily mediate the replication of latent viral episomes and their partitioning to daughter cells during mitosis. On the other hand, lytic proteins such as HHV-6A IE2 and HCMV IE2 of the beta herpesvirus subfamily act as transcriptional activators of early or late viral promoters.
Based on the crystal structures, the DNA-binding domains (DBDs) of LANA-1, EBNA-1, and HHV-6A IE2 share significant structural homology. AlphaFold models of HCMV IE2 predict a similar fold structure which indicates that DBD is conserved among herpesvirus families. In LANA-1, there are two caspase I and III cleavage sites. We hypothesize that a physiologically relevant domain is formed in the C-terminal domain (CTD) after being cleaved by caspase III, which we refer to as the extended CTD (eCTD).
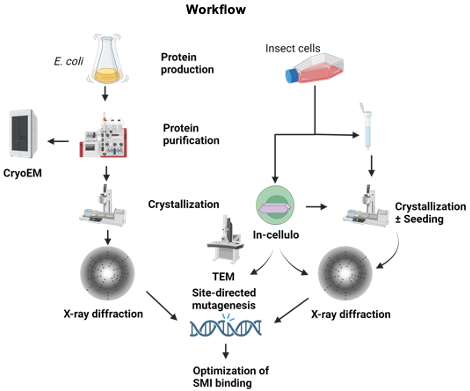
The aim of the project is the structural and functional characterization of LANA DBD, eCTD, and HCMV-IE2 CTD alone and in complex with the viral DNA and small molecular inhibitors (SMIs) to develop innovative targeted therapy. We plan to employ conventional X-ray crystallography, in cellulo crystallization techniques, and cryo-EM for structure elucidation
C1
References
- Schulz T. F. et al. Current Opinion in Virology, https://doi.org/10.1016/j.coviro.2023.101336 (2023).
- Nishimura et al. Journal of Virology, https://doi.org/10.1128/JVI.01121-17 (2017).
- Hellert, J. et al. Proceedings of the National Academy of Sciences of the United States of America, https://doi.org/10.1073/pnas.1421804112 (2015).
- Bochkarev A. et al. Cell, https://doi.org/10.1016/0092-8674(95)90232-5 (1995).
- Berwanger, A. et al. Journal of Medicinal Chemistry, https://doi.org/10.1021/acs.jmedchem.3c00990 (2023).
- Kirsch, P. et al. Journal of Medicinal Chemistry, https://doi.org/10.1021/acs.jmedchem.8b01827 (2019).
- Kirsch, P. et al. European Journal of Medicinal Chemistry, https://doi.org/10.1016/j.ejmech.2020.112525 (2020).
- Messick, T. E. et al. Science Translational Medicine, https://doi.org/10.1126/scitranslmed.aau5612 (2019).
- Redecke, L. et al. Science, https://doi.org/10.1126/science.1229663 (2013).
- Nass, K. et al. Nat Commun, https://doi.org/10.1038/s41467-020-14484-w (2020).
