Principal Investigator
Prof. Dr. Kay Grünewald
CSSB Center for Structural Systems Biology
Universität Hamburg/Leibniz Institute of Virology


B1
PhD candidate
Julia Nentwig

B1
Project Summary
High-resolution structure and conformational flexibility of Herpes simplex virus 1 (HSV-1) fusion protein gB.
Herpes simplex virus-1 (HSV-1) virions are covered with hundreds of glycoproteins of more than 12 different kinds [1]. The fusion machinery of HSV-1, responsible for mediating fusion of the viral membrane with the plasma or endosomal membrane, consists of four essential glycoproteins, including glycoprotein B (gB) which is the bona fide fusion protein. During the fusion process gB changes its conformation from a pre-fusion to a post-fusion state. While the post-fusion conformation is highly stable and has been structurally resolved already in 2006 using X-ray crystallography, the pre-fusion conformation of gB eluded scientist for a long time due to its metastability, which is inherent to many pre-fusion forms of other viral fusion proteins as well. Recently, the pre-fusion conformation was solved using sub-volume averaging from cryo-electron tomography (cryo-ET) data after insertion of a stabilizing mutation [2]. The pre-fusion conformation of viral fusion proteins can be an important target for prevention and treatment of viral infections. Furthermore, solving the pre-fusion structure was the first step in understanding the fusion mechanism in more detail.
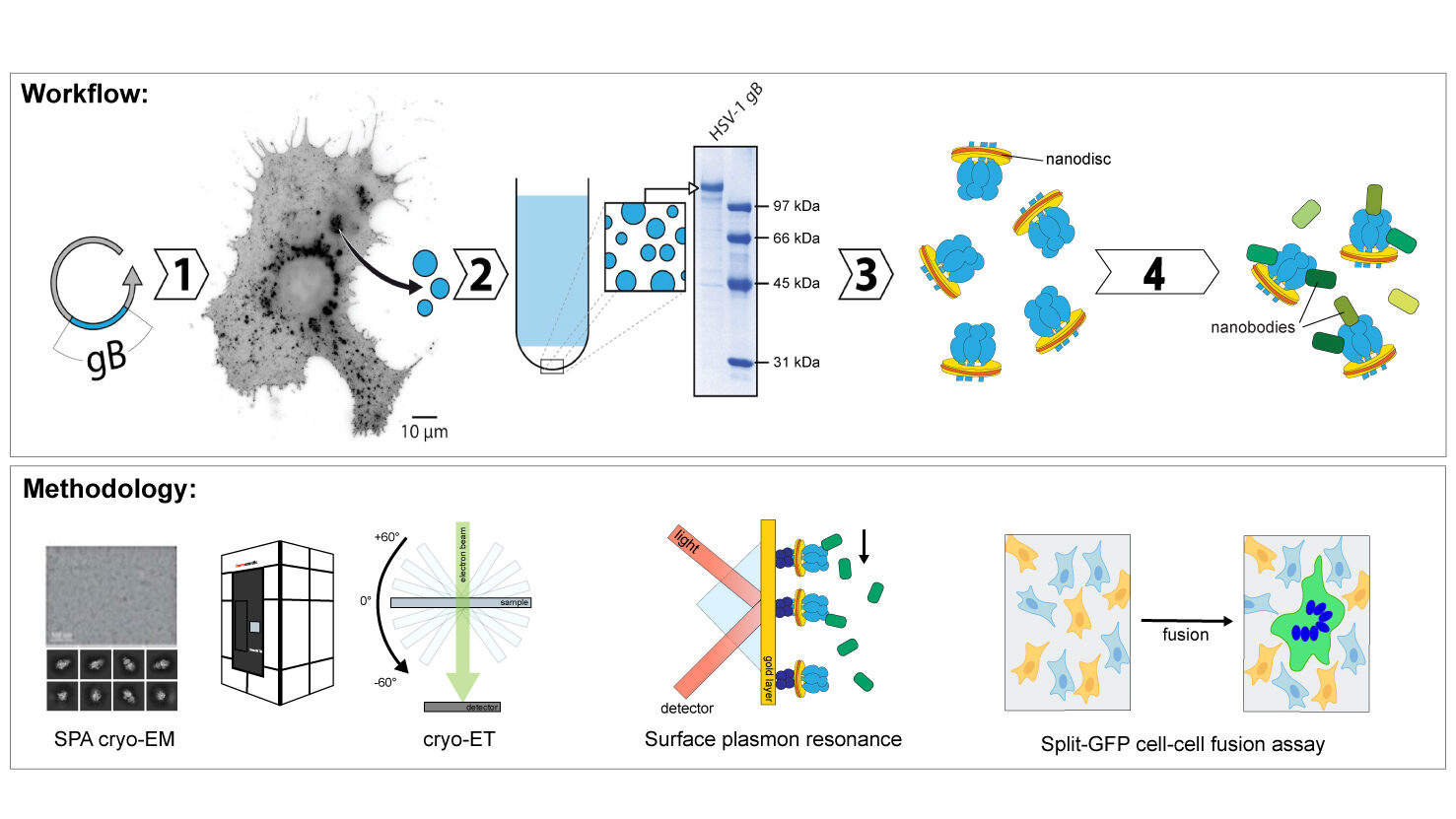
Nanobodies are camelid single domain-only antibodies that are much smaller than regular antibodies, which makes them a perfect tool to study function and structure of proteins. Moreover, they are easily produced by recombinant expression in E. coli. Nanobodies can be used for biological research, bioimaging, disease diagnosis or as targeted therapeutics [3,4].
The goal of this project is to characterise conformation-specific nanobodies against HSV-1 gB with the prospect of using them as a tool for structural and functional studies. Therefore, we employ biophysical, cell biological and structural techniques to determine their binding affinities, functional impact and binding sites on gB. Additionally, nanobodies will also be screened for cross-species reactivity with gB of other herpes viruses.
B1
References
- Grünewald K, Desai P, Winkler DC, et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003 Nov 21;302(5649):1396-8. doi: 10.1126/science.1090284
- Vollmer B, Pražák V, Vasishtan D, et al. The prefusion structure of herpes simplex virus glycoprotein B. Sci Adv. 2020 Sep 25;6(39):eabc1726. doi: 10.1126/sciadv.abc1726
- Frecot DI, Froehlich T, Rothbauer U. 30 years of nanobodies – an ongoing success story of small binders in biological research. J Cell Sci. 2023 Nov 1;136(21):jcs261395. doi: 10.1242/jcs.261395
- Matthys A, Saelens X. Promises and challenges of single-domain antibodies to control influenza. Antiviral Res. 2024 Feb;222:105807. doi: 10.1016/j.antiviral.2024.105807
